自己投資としてチャレンジしている内容を Blog で公開しています。
今回は Apache Spark の環境で利用できる pyspark spark-xml を使い、COVID-19 に関わる SARS-CoV-2 の Spile 蛋白質の情報を UniProt にある xml ファイルから抽出する方法を紹介します。
————————————
▼1. Covid-19 Spike 蛋白質
————————————
COVID-19 (Coronavirus Disease 2019) は、SARS-CoV-2 (Sever Acute Respiratory Syndrome Corona Virus 2) を病原体としています。この SARS-CoV-2 の構造タンパク質の内、Spike Glycoprotein はウィルス粒子が細胞に侵入する際に利用される接着部分となります。この S protein はワクチン開発のターゲットとして重要な部分として知られています。今回はこの Spike Glycoprotein である UniProtKB – P0DTC2 (SPIKE_SARS2) “P0DTC2.xml” から特徴を抽出します。
SARS-CoV-2 の構造タンパク質には以下が含まれます。
- Envelope Protein (E)
- Spike もしくは Surface Glycoprotein (S)
- Membrance Protein (M)
- Nucleocapsid Protein (N)
– 新型コロナウイルス感染症の世界的流行 (2019年-) – Wikipedia
– S surface glycoprotein [Severe acute respiratory syndrome coronavirus 2] – Gene – NCBI (nih.gov)
————————————
▼2. XML ファイルのデータの読み取り
————————————
Apache Spark 3 の環境で XML ファイルのデータを読み取るために、以下の spark-xml のライブラリーを利用します。
例) com.databricks:spark-xml_2.12:0.14.0 の場合、2022年 5 月の時点で最新の version は 2.12 の 0.14.0 となります。com.databricks : spark-xml_2.12 – Maven Central Repository Search
————————————
▼3. 事前準備
————————————
3-1. Spark-xml を利用可能な環境を以下を参考に構築します。
Spark-xml を使ったXML データの読み取り、XML ファイル作成 No.50 – 2022/05
2-2. Databricks の spark-xml の jar をダウンロードします。
spark-xml_2.12-0.14.0.jar の場合 com.databricks : spark-xml_2.12 : 0.14.0 – Maven Central Repository Search
例) wget https://repo1.maven.org/maven2/com/databricks/spark-xml_2.12/0.14.0/spark-xml_2.12-0.14.0.jar3-3. XML document と Java Object をマッピングする JAXB の jar をダウンロードします。
jaxb-core-3.0.0-M2.jar の場合 https://repo1.maven.org/maven2/com/sun/xml/bind/jaxb-core/3.0.0-M2/
例) wget https://repo1.maven.org/maven2/com/sun/xml/bind/jaxb-core/3.0.0-M2/jaxb-core-3.0.0-M2.jar3-4. サンプルの P0DTC2.xml ファイルをダウンロードします。
https://www.uniprot.org/uniprot/P0DTC2.xml
S – Spike glycoprotein precursor – Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) – S gene & protein (uniprot.org)
3-5. ダウンロードした P0DTC2.xml を、事前に作成しておいた HDFS のディレクトリー appbox に置きます。
(例)
hdfs dfs -put ./P0DTC2.xml appbox————————————
▼4. Spike Glycoprotein に含まれる “特徴” と “アミノ酸配列の場所” をまとめる
————————————
以下の順に進めます。
———————
4-1. spark-xml の jar ファイル及び、XML document と Java Object をマッピングする JAXB の jar を指定して pyspark を起動します。
4-2. DataFrame を作成するために SparkSession を作成します。
4-3. P0DTC2.xml ファイルからデータ部分読み取り DataFrame に入れます。
4-4. さらに列を絞り、特徴とその場所を抽出します。
4-5. アミノ酸の配列の場所がきれいに表示できていないことを確認します。
4-6. Location 列のStructType の列を、通常の列に変換します。
4-6. Spark UI で Spark Job の実行履歴を確認します。
———————
4-1. spark-xml の jar ファイル及び、XML document と Java Object をマッピングする JAXB の jar を指定して pyspark を起動します。
$ pyspark --jars spark-xml_2.12-0.14.0.jar,jaxb-core-3.0.0-M2.jar
Python 3.8.10 (default, Sep 28 2021, 16:10:42)
[GCC 9.3.0] on linux
Type "help", "copyright", "credits" or "license" for more information.
2021-10-19 22:26:57,756 WARN util.NativeCodeLoader: Unable to load native-hadoop library for your platform... using builtin-java classes where applicable
Setting default log level to "WARN".
To adjust logging level use sc.setLogLevel(newLevel). For SparkR, use setLogLevel(newLevel).
Welcome to
____ __
/ __/__ ___ _____/ /__
_\ \/ _ \/ _ `/ __/ '_/
/__ / .__/\_,_/_/ /_/\_\ version 3.1.2
/_/
Using Python version 3.8.10 (default, Sep 28 2021 16:10:42)
Spark context Web UI available at http://xxxxx:4040
Spark context available as 'sc' (master = local[*], app id = local-1634650023016).
SparkSession available as 'spark'.
>>> 4-2. DataFrame を作成するために pyspark.sql.SparkSession をインポートし、SparkSession を作成します。
>>> from pyspark.sql import SparkSession
>>> spark = SparkSession.builder.getOrCreate()4-3. P0DTC2.xml ファイルからデータを読み取り DataFrame に入れます。
>>> df = spark.read.format('xml').options(rowTag='feature').load('appbox/P0DTC2.xml')
>>> df.show(5)
+------------------+---------+--------------+--------------+--------------------+--------+---------+
| _description|_evidence| _id| _type| location|original|variation|
+------------------+---------+--------------+--------------+--------------------+--------+---------+
| null| 35| null|signal peptide|{{null, 1}, {null...| null| null|
|Spike glycoprotein| null|PRO_0000449646| chain|{{null, 14}, {nul...| null| null|
| Spike protein S1| 2|PRO_0000449647| chain|{{null, 14}, {nul...| null| null|
| Spike protein S2| 2|PRO_0000449648| chain|{{null, 686}, {nu...| null| null|
| Spike protein S2'| 2|PRO_0000449649| chain|{{null, 816}, {nu...| null| null|
+------------------+---------+--------------+--------------+--------------------+--------+---------+
only showing top 5 rows4-4. さらに列を絞り、特徴とその場所を抽出します。
>>> selectdpro2 = df.select("_description","_id","_type","location")
>>> selectdpro2.printSchema()
>>> selectdpro2.printSchema()
root
|-- _description: string (nullable = true)
|-- _id: string (nullable = true)
|-- _type: string (nullable = true)
|-- location: struct (nullable = true)
| |-- begin: struct (nullable = true)
| | |-- _VALUE: string (nullable = true)
| | |-- _position: long (nullable = true)
| |-- end: struct (nullable = true)
| | |-- _VALUE: string (nullable = true)
| | |-- _position: long (nullable = true)
| |-- position: struct (nullable = true)
| | |-- _VALUE: string (nullable = true)
| | |-- _position: long (nullable = true)
>>> selectdpro2.show()
+--------------------+--------------+--------------------+--------------------+
| _description| _id| _type| location|
+--------------------+--------------+--------------------+--------------------+
| null| null| signal peptide|{{null, 1}, {null...|
| Spike glycoprotein|PRO_0000449646| chain|{{null, 14}, {nul...|
| Spike protein S1|PRO_0000449647| chain|{{null, 14}, {nul...|
| Spike protein S2|PRO_0000449648| chain|{{null, 686}, {nu...|
| Spike protein S2'|PRO_0000449649| chain|{{null, 816}, {nu...|
| Extracellular| null| topological domain|{{null, 14}, {nul...|
| Helical| null|transmembrane region|{{null, 1214}, {n...|
| Cytoplasmic| null| topological domain|{{null, 1235}, {n...|
| BetaCoV S1-NTD| null| domain|{{null, 14}, {nul...|
| BetaCoV S1-CTD| null| domain|{{null, 334}, {nu...|
|Putative superant...| null| region of interest|{{null, 280}, {nu...|
|Receptor-binding ...| null| region of interest|{{null, 319}, {nu...|
|Integrin-binding ...| null| region of interest|{{null, 403}, {nu...|
|Receptor-binding ...| null| region of interest|{{null, 437}, {nu...|
|Immunodominant HL...| null| region of interest|{{null, 448}, {nu...|
|Putative superant...| null| region of interest|{{null, 681}, {nu...|
| Fusion peptide 1| null| region of interest|{{null, 816}, {nu...|
| Fusion peptide 2| null| region of interest|{{null, 835}, {nu...|
| Heptad repeat 1| null| region of interest|{{null, 920}, {nu...|
| Heptad repeat 2| null| region of interest|{{null, 1163}, {n...|
+--------------------+--------------+--------------------+--------------------+
only showing top 20 rows
>>> selectdpro2.filter("_id is not null").show(10,False)
+------------------+--------------+-----+---------------------------------+
|_description |_id |_type|location |
+------------------+--------------+-----+---------------------------------+
|Spike glycoprotein|PRO_0000449646|chain|{{null, 14}, {null, 1273}, null} |
|Spike protein S1 |PRO_0000449647|chain|{{null, 14}, {null, 685}, null} |
|Spike protein S2 |PRO_0000449648|chain|{{null, 686}, {null, 1273}, null}|
|Spike protein S2' |PRO_0000449649|chain|{{null, 816}, {null, 1273}, null}|
+------------------+--------------+-----+---------------------------------+
>>> selectdpro2.show(20,False)
+---------------------------------------------------------------------+--------------+--------------------+----------------------------------+
|_description |_id |_type |location |
+---------------------------------------------------------------------+--------------+--------------------+----------------------------------+
|null |null |signal peptide |{{null, 1}, {null, 13}, null} |
|Spike glycoprotein |PRO_0000449646|chain |{{null, 14}, {null, 1273}, null} |
|Spike protein S1 |PRO_0000449647|chain |{{null, 14}, {null, 685}, null} |
|Spike protein S2 |PRO_0000449648|chain |{{null, 686}, {null, 1273}, null} |
|Spike protein S2' |PRO_0000449649|chain |{{null, 816}, {null, 1273}, null} |
|Extracellular |null |topological domain |{{null, 14}, {null, 1213}, null} |
|Helical |null |transmembrane region|{{null, 1214}, {null, 1234}, null}|
|Cytoplasmic |null |topological domain |{{null, 1235}, {null, 1273}, null}|
|BetaCoV S1-NTD |null |domain |{{null, 14}, {null, 303}, null} |
|BetaCoV S1-CTD |null |domain |{{null, 334}, {null, 527}, null} |
|Putative superantigen; may bind T-cell receptor alpha/TRAC |null |region of interest |{{null, 280}, {null, 301}, null} |
|Receptor-binding domain (RBD) |null |region of interest |{{null, 319}, {null, 541}, null} |
|Integrin-binding motif; |null |region of interest |{{null, 403}, {null, 405}, null} |
|Receptor-binding motif; binding to human ACE2 |null |region of interest |{{null, 437}, {null, 508}, null} |
|Immunodominant HLA epitope recognized by the CD8+; called NF9 peptide|null |region of interest |{{null, 448}, {null, 456}, null} |
|Putative superantigen; may bind T-cell receptor beta/TRBC1 |null |region of interest |{{null, 681}, {null, 684}, null} |
|Fusion peptide 1 |null |region of interest |{{null, 816}, {null, 837}, null} |
|Fusion peptide 2 |null |region of interest |{{null, 835}, {null, 855}, null} |
|Heptad repeat 1 |null |region of interest |{{null, 920}, {null, 970}, null} |
|Heptad repeat 2 |null |region of interest |{{null, 1163}, {null, 1202}, null}|
+---------------------------------------------------------------------+--------------+--------------------+----------------------------------+
only showing top 20 rows4-5. アミノ酸の配列の場所がきれいに表示できていないことを確認します。
4-4 で列を絞り、特徴とその場所を抽出することができました。ただ、 Spike glycoprotein の場所が以下のように表示されています。Begin が 14 で End が 1273 を示しています。
{{null, 14}, {null, 1273}, null}
この表示の理由は、4-4 で selectdrop2 の dataframe の schema を表示した際確認できたように、StructType の列を含んでいるためです。この点改善するため、以降では、この StructType の列を、通常の列に変換します。
>>> selectdpro2.printSchema()
root
|-- _description: string (nullable = true)
|-- _id: string (nullable = true)
|-- _type: string (nullable = true)
|-- location: struct (nullable = true)
| |-- begin: struct (nullable = true)
| | |-- _VALUE: string (nullable = true)
| | |-- _position: long (nullable = true)
| |-- end: struct (nullable = true)
| | |-- _VALUE: string (nullable = true)
| | |-- _position: long (nullable = true)
| |-- position: struct (nullable = true)
| | |-- _VALUE: string (nullable = true)
| | |-- _position: long (nullable = true)4-6. Location 列のStructType の列を、通常の列に変換します。
最終的には、Spike glycoprotein の場所の表示が以下のように改善されました。
(変更前)
{{null, 14}, {null, 1273}, null}
(変更後)
14| 1273|
Ref: How to Convert Struct type to Columns in Spark – Spark by {Examples} (sparkbyexamples.com)
>>> df = spark.read.format('xml').options(rowTag='feature').load('appbox/P0DTC2.xml')
>>> df.printSchema()
root
|-- _description: string (nullable = true)
|-- _evidence: string (nullable = true)
|-- _id: string (nullable = true)
|-- _type: string (nullable = true)
|-- location: struct (nullable = true)
| |-- begin: struct (nullable = true)
| | |-- _VALUE: string (nullable = true)
| | |-- _position: long (nullable = true)
| |-- end: struct (nullable = true)
| | |-- _VALUE: string (nullable = true)
| | |-- _position: long (nullable = true)
| |-- position: struct (nullable = true)
| | |-- _VALUE: string (nullable = true)
| | |-- _position: long (nullable = true)
|-- original: string (nullable = true)
|-- variation: array (nullable = true)
| |-- element: string (containsNull = true)
>>> df2 = df.select('_description','_evidence','_id','_type','location.begin.*','location.end.*','location.position.*','original')
>>> df2Flatten = df2.toDF("_description","_evidence","_id","_type","location_begin_value","location_begin_position","location_end_value","location_end_position","location_position_value","location_position_position","original")
>>> df2Flatten.printSchema()
root
|-- _description: string (nullable = true)
|-- _evidence: string (nullable = true)
|-- _id: string (nullable = true)
|-- _type: string (nullable = true)
|-- location_begin_value: string (nullable = true)
|-- location_begin_position: long (nullable = true)
|-- location_end_value: string (nullable = true)
|-- location_end_position: long (nullable = true)
|-- location_position_value: string (nullable = true)
|-- location_position_position: long (nullable = true)
|-- original: string (nullable = true)
>>> df2Flatten2 =df2Flatten.select("_description","_id","_type","location_begin_position","location_end_position")
>>> df2Flatten2.filter("_id is not null").show()
+------------------+--------------+-----+-----------------------+---------------------+
| _description| _id|_type|location_begin_position|location_end_position|
+------------------+--------------+-----+-----------------------+---------------------+
|Spike glycoprotein|PRO_0000449646|chain| 14| 1273|
| Spike protein S1|PRO_0000449647|chain| 14| 685|
| Spike protein S2|PRO_0000449648|chain| 686| 1273|
| Spike protein S2'|PRO_0000449649|chain| 816| 1273|
+------------------+--------------+-----+-----------------------+---------------------+
>>> df2Flatten2.show(30,False)
+---------------------------------------------------------------------+--------------+---------------------------+-----------------------+---------------------+
|_description |_id |_type |location_begin_position|location_end_position|
+---------------------------------------------------------------------+--------------+---------------------------+-----------------------+---------------------+
|null |null |signal peptide |1 |13 |
|Spike glycoprotein |PRO_0000449646|chain |14 |1273 |
|Spike protein S1 |PRO_0000449647|chain |14 |685 |
|Spike protein S2 |PRO_0000449648|chain |686 |1273 |
|Spike protein S2' |PRO_0000449649|chain |816 |1273 |
|Extracellular |null |topological domain |14 |1213 |
|Helical |null |transmembrane region |1214 |1234 |
|Cytoplasmic |null |topological domain |1235 |1273 |
|BetaCoV S1-NTD |null |domain |14 |303 |
|BetaCoV S1-CTD |null |domain |334 |527 |
|Putative superantigen; may bind T-cell receptor alpha/TRAC |null |region of interest |280 |301 |
|Receptor-binding domain (RBD) |null |region of interest |319 |541 |
|Integrin-binding motif; |null |region of interest |403 |405 |
|Receptor-binding motif; binding to human ACE2 |null |region of interest |437 |508 |
|Immunodominant HLA epitope recognized by the CD8+; called NF9 peptide|null |region of interest |448 |456 |
|Putative superantigen; may bind T-cell receptor beta/TRBC1 |null |region of interest |681 |684 |
|Fusion peptide 1 |null |region of interest |816 |837 |
|Fusion peptide 2 |null |region of interest |835 |855 |
|Heptad repeat 1 |null |region of interest |920 |970 |
|Heptad repeat 2 |null |region of interest |1163 |1202 |
|null |null |coiled-coil region |949 |993 |
|null |null |coiled-coil region |1176 |1203 |
|Binding to host endocytosis trafficking protein SNX27 |null |short sequence motif |1237 |1241 |
|Diacidic ER export motif (host COPII) |null |short sequence motif |1257 |1262 |
|Binding to host plasma membrane localising/FERM domain proteins |null |short sequence motif |1261 |1267 |
|KxHxx, ER retrieval signal (COPI) |null |short sequence motif |1269 |1273 |
|Cleavage; by TMPRSS2 or furin |null |site |685 |686 |
|Cleavage; by host |null |site |815 |816 |
|S-palmitoyl cysteine; by host ZDHHC20 |null |lipid moiety-binding region|null |null |
|S-palmitoyl cysteine; by host ZDHHC20 |null |lipid moiety-binding region|null |null |
+---------------------------------------------------------------------+--------------+---------------------------+-----------------------+---------------------+
only showing top 30 rows4-7. feature を抽出する以外に、この蛋白質 P0DTC2 に関わる論文 reference を抽出することもできます。
>>> dfref = spark.read.format('xml').options(rowTag='reference').load('appbox/P0DTC2.xml')
>>> dfref.printSchema()
root
|-- _evidence: string (nullable = true)
|-- _key: long (nullable = true)
|-- citation: struct (nullable = true)
| |-- _date: long (nullable = true)
| |-- _first: string (nullable = true)
| |-- _last: string (nullable = true)
| |-- _name: string (nullable = true)
| |-- _type: string (nullable = true)
| |-- _volume: long (nullable = true)
| |-- authorList: struct (nullable = true)
| | |-- consortium: struct (nullable = true)
| | | |-- _VALUE: string (nullable = true)
| | | |-- _name: string (nullable = true)
| | |-- person: array (nullable = true)
| | | |-- element: struct (containsNull = true)
| | | | |-- _VALUE: string (nullable = true)
| | | | |-- _name: string (nullable = true)
| |-- dbReference: array (nullable = true)
| | |-- element: struct (containsNull = true)
| | | |-- _VALUE: string (nullable = true)
| | | |-- _id: string (nullable = true)
| | | |-- _type: string (nullable = true)
| |-- title: string (nullable = true)
|-- scope: array (nullable = true)
| |-- element: string (containsNull = true)
|-- source: struct (nullable = true)
| |-- strain: array (nullable = true)
| | |-- element: string (containsNull = true)
>>> dfref.show()4-8. 最後に、Spark UI で Spark のジョブ実行履歴を確認します。
- クラスタの状況を確認 http://localhost:8080/cluster/
- ジョブの実行履歴を確認 http://localhost:4040/job/
1. クラスタの状況を確認 http://localhost:8080/cluster/
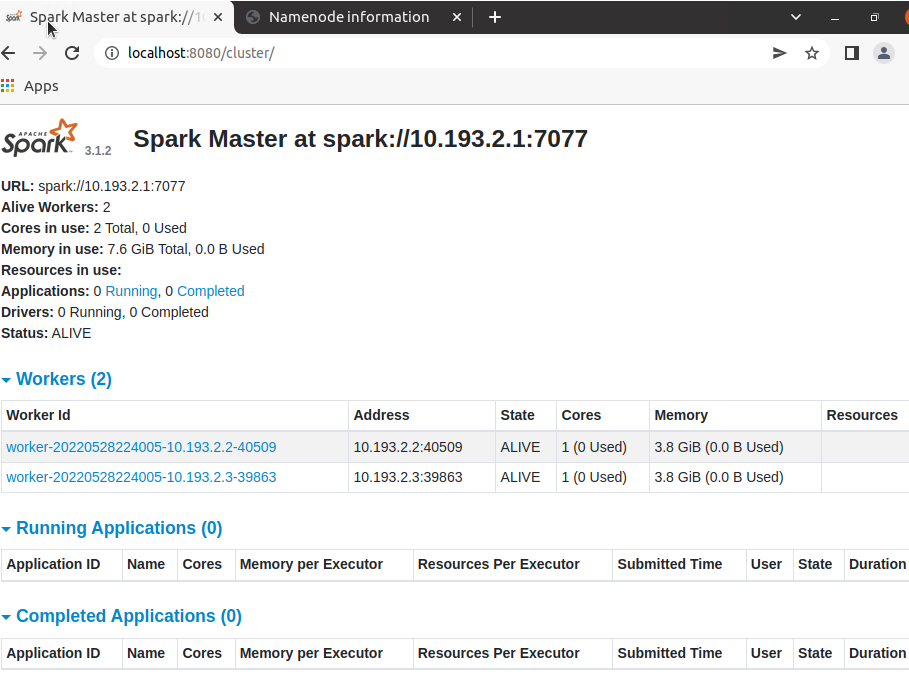
2. ジョブの実行履歴を確認 http://localhost:4040/job/

————————————
▼5. 参考情報
————————————
(1) databricks/spark-xml: XML data source for Spark SQL and DataFrames (github.com)
(2) S – Spike glycoprotein precursor – Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) – S gene & protein (uniprot.org)
(3) How to Convert Struct type to Columns in Spark – Spark by {Examples} (sparkbyexamples.com)
以上です。参考になりましたら幸いです。